Analysis of the UK Biobank data
Pooled analysis of all populations
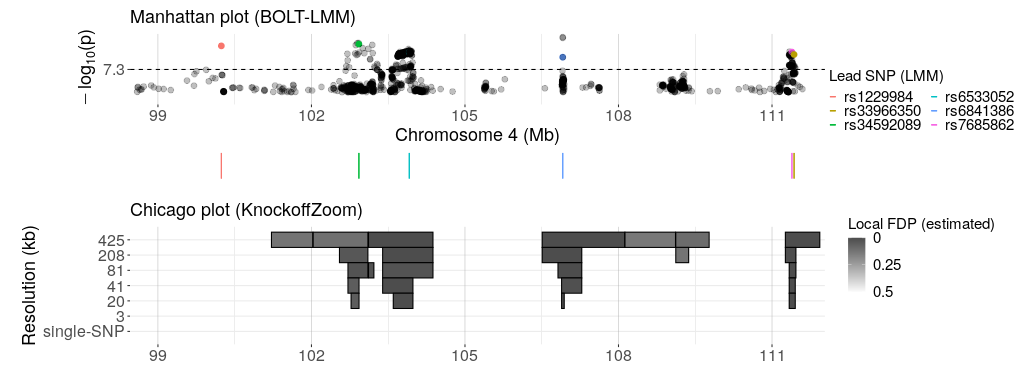
We apply KnockoffGWAS to several phenotypes in the UK Biobank data and obtain many new findings.
To learn more about this analysis and the method behind it, read the following accompanying paper.
FDR control in GWAS with population structure
M. Sesia, S. Bates, E. Candès, J. Marchini, C. Sabatti
preprint at bioRxiv; doi:10.1101/2020.08.04.236703
To produce more plots like the one below and explore our results interactively, visit: https://sesia.shinyapps.io/knockoffgwas/.
Download the complete list of multi-resolution KnockoffGWAS discoveries for each trait.
Download the complete list of KnockoffGWAS test statistics at each resolution.
Multi-population analysis searching for robust associations
The following paper applies KnockoffGWAS to the UK Biobank data to discover genetic associations that are invariant across populations with different ancestries.
Searching for consistent associations with a multi-environment knockoff filter
S. Li, M. Sesia, Y. Romano, E. Candès, C Sabatti
preprint at arXiv; https://arxiv.org/abs/2106.04118
Download the complete list of multi-resolution KnockoffGWAS invariant discoveries for each trait.
Transfer learning analysis across populations
The following paper applies KnockoffGWAS to the UK Biobank data to discover genetic associations leveraging prior information in external data sets collected from different populations or measuring related outcomes.
Transfer learning in genome-wide association studies with knockoffs
S. Li, Z. Ren, C. Sabatti, M. Sesia
preprint at arXiv; https://arxiv.org/abs/2108.08813
Download the complete list of multi-resolution KnockoffGWAS transfer learning discoveries.