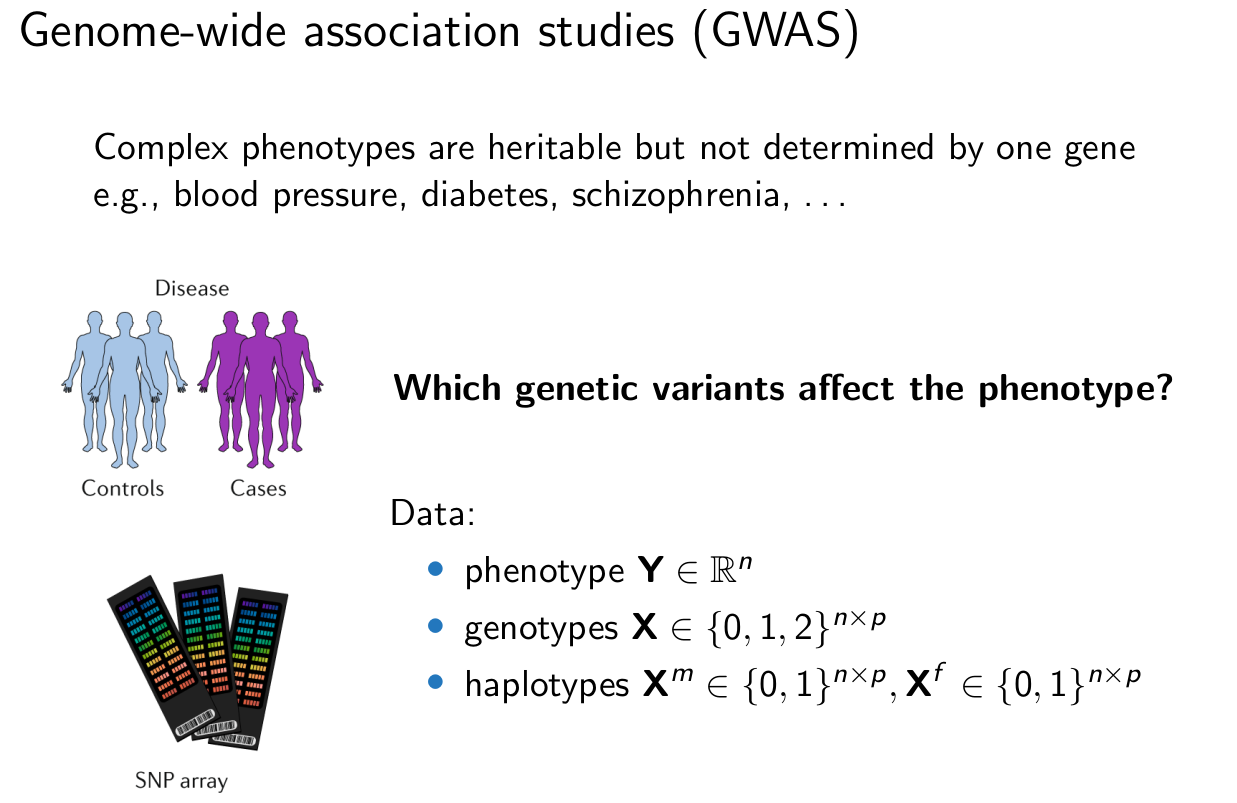
Traditional approach: testing marginal associations¶
Methods: univariate regression, permutations, linear mixed models.
\begin{align*} \text{marginal association} \quad & \neq \quad \text{causation} \\ Y {\; \not\!\perp\!\!\!\perp \;} X_j \quad & \qquad X_j \rightarrow Y \end{align*}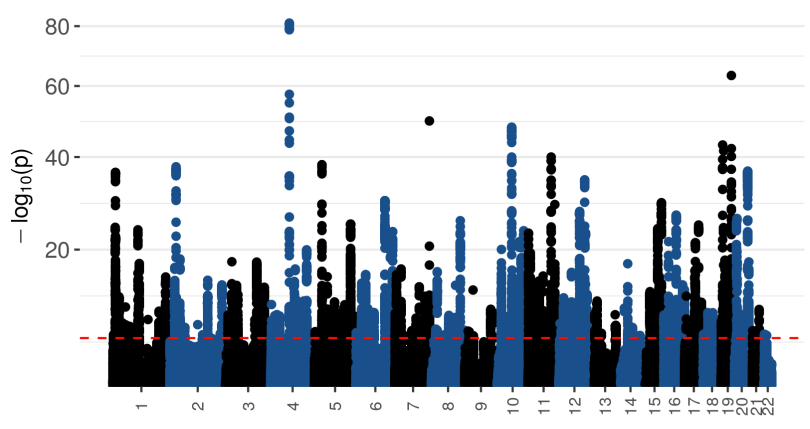
Variants on same chromosome are not independent of each other (linkage disequilibrium, population structure, familial relatedness).
Testing conditional associations¶
Partition the variables into (contiguous) groups.
\begin{align*} \underbrace{X_1, X_2, X_3}_{G_1},\underbrace{X_4, X_5}_{G_2}, \underbrace{X_6, X_7}_{G_3}, \underbrace{X_8, X_9, X_{10}}_{G_4} \end{align*}
Conditional hypotheses: \begin{align*} \mathcal{H}_{0,g} : Y {\; \perp\!\!\!\perp \;} X_g \mid X_{-g}. \end{align*}
- Larger groups: easier to reject the null
- Smaller groups: more informative discoveries
Control the false discovery rate: the expected proportion of spurious discoveries.
Model-X framework: \begin{align*} (X^i,Y^i) \overset{}{\sim} P_{XY} = P_X \cdot P_{Y \mid X}, \qquad X^i \in \mathbb{R}^p, Y^i \in \mathbb{R}. \end{align*} Model $P_X$, make inferences about $P_{Y \mid X}$.
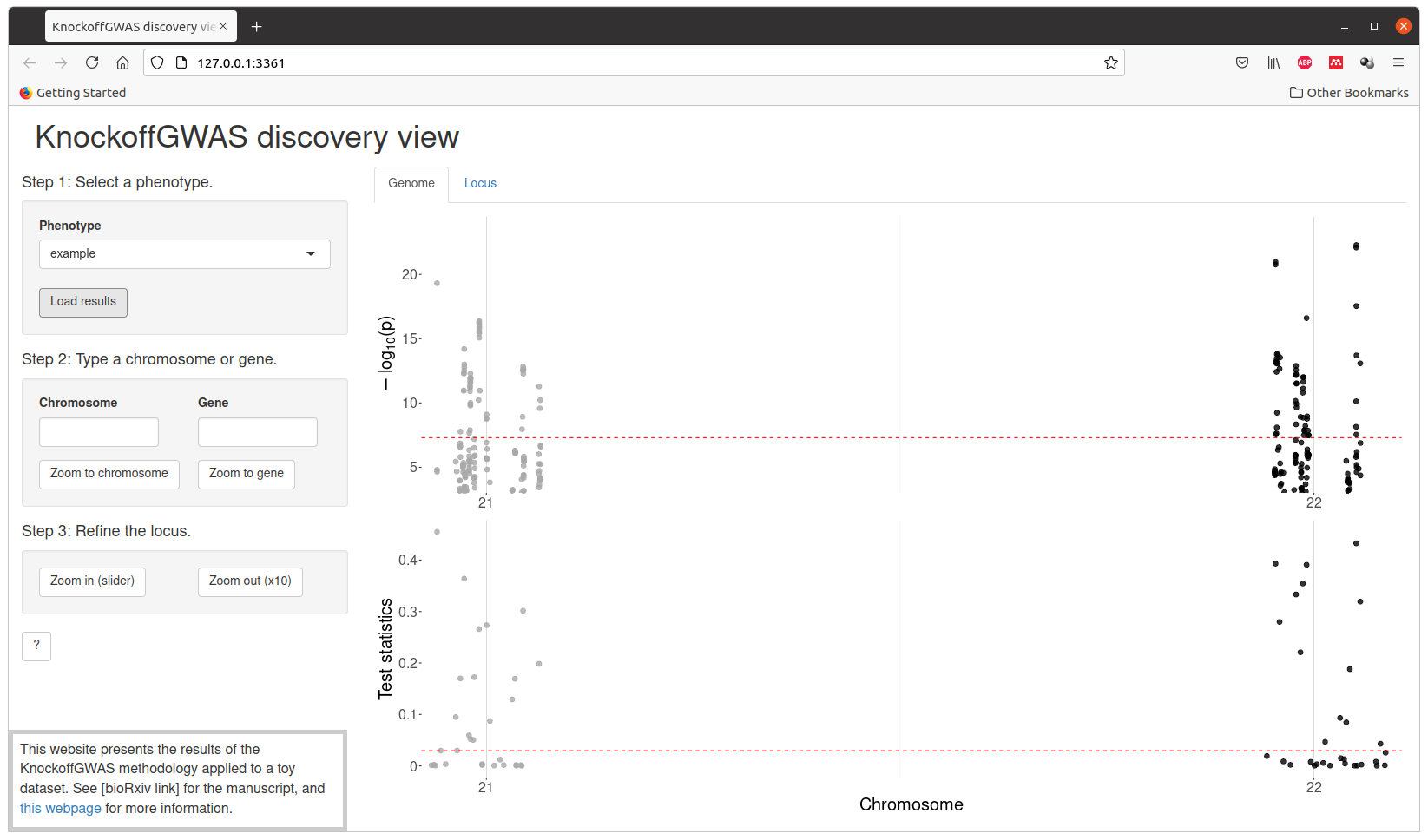
Preview of discoveries (simulated phenotype)¶
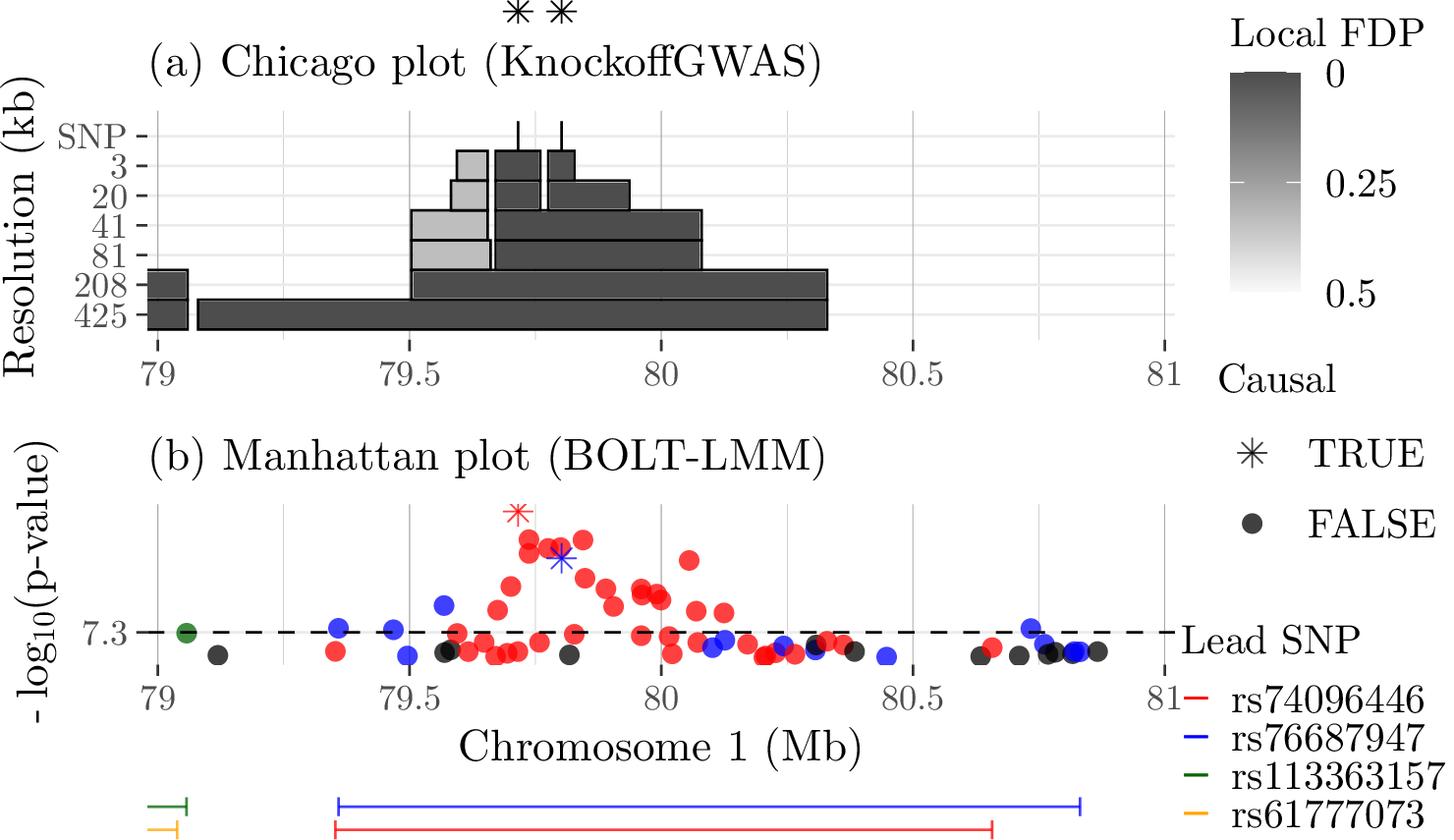
Genotypes and knockoffs¶
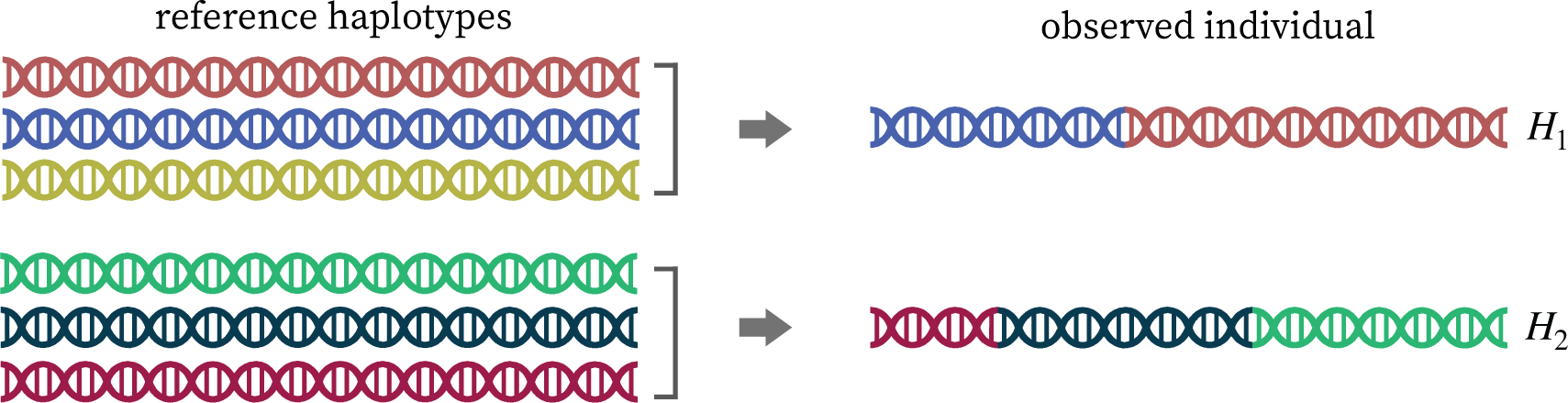
Knockoff are synthetic variables that "look like", but are not identical to, the genotypes.
Knockoffs preserve allele frequencies, linkage disequilibrium, and population structure.
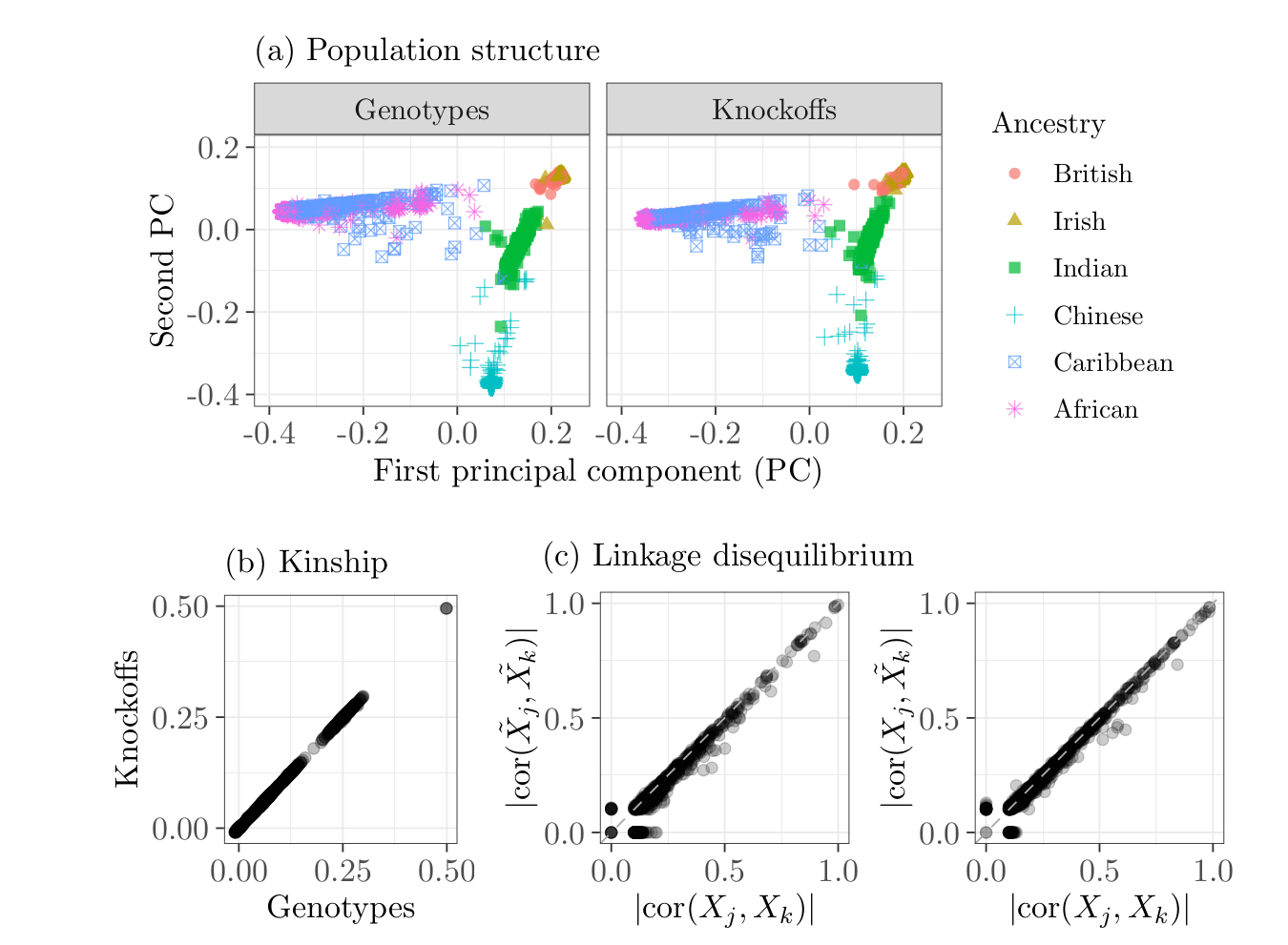
Knockoffs as negative control variables¶
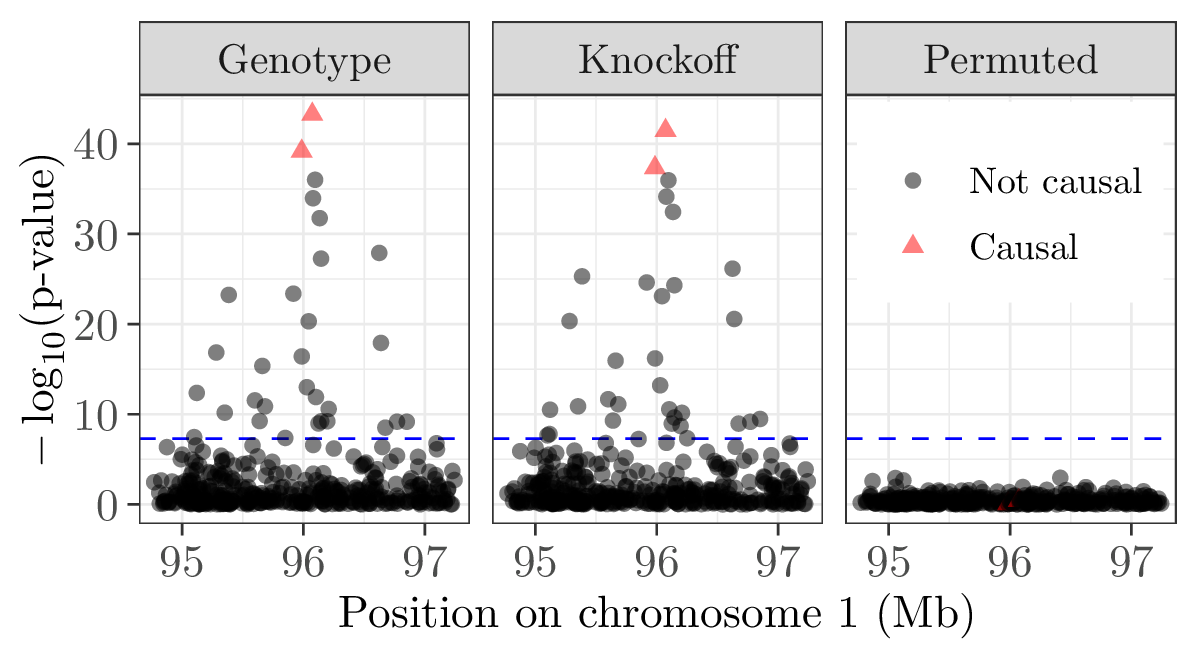
Main strength of knockoffs: they can be used as negative control variables with any model, including complicated multivariate and machine learning models.
Tutorial¶
A toy genetic dataset containing 1000 artificial samples typed at 2000 loci (divided between chromosome 21 and 22) is available from the software repository.
Download the repository, enter the main directory, and compile the snpknock2 C++ program by typing in your terminal:
cd snpknock2
make
cd ..After a successful compilation, you can execute the script analyze.sh.
./analyze.shThis will first verify whether the system dependencies and then carry out an entire association analysis.
This analysis consists of 4 main modules.
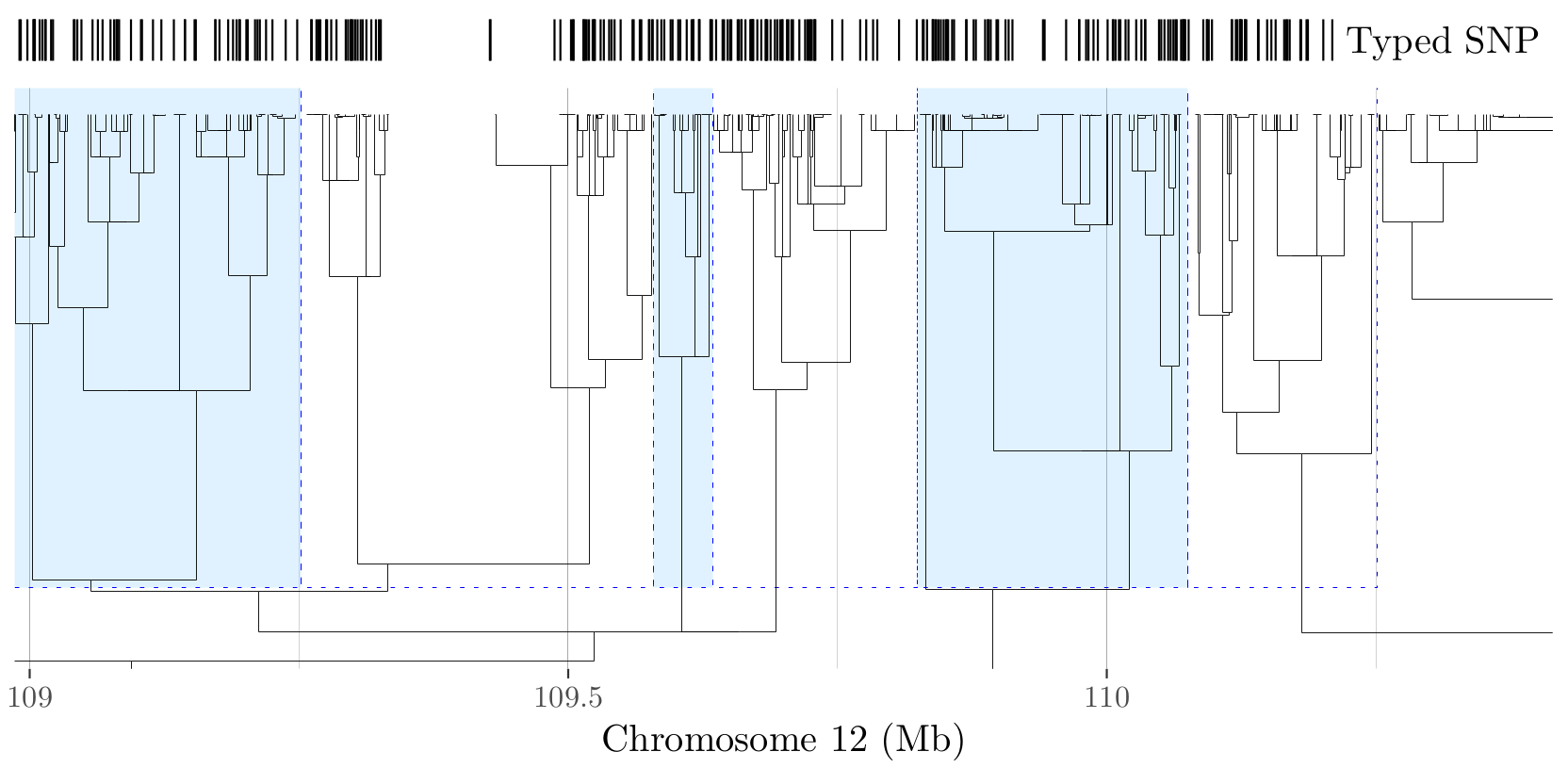
Module 1: partition the genome¶
Partition the available SNPs into contiguous groups at different levels of resolution, based on the genetic distance information.
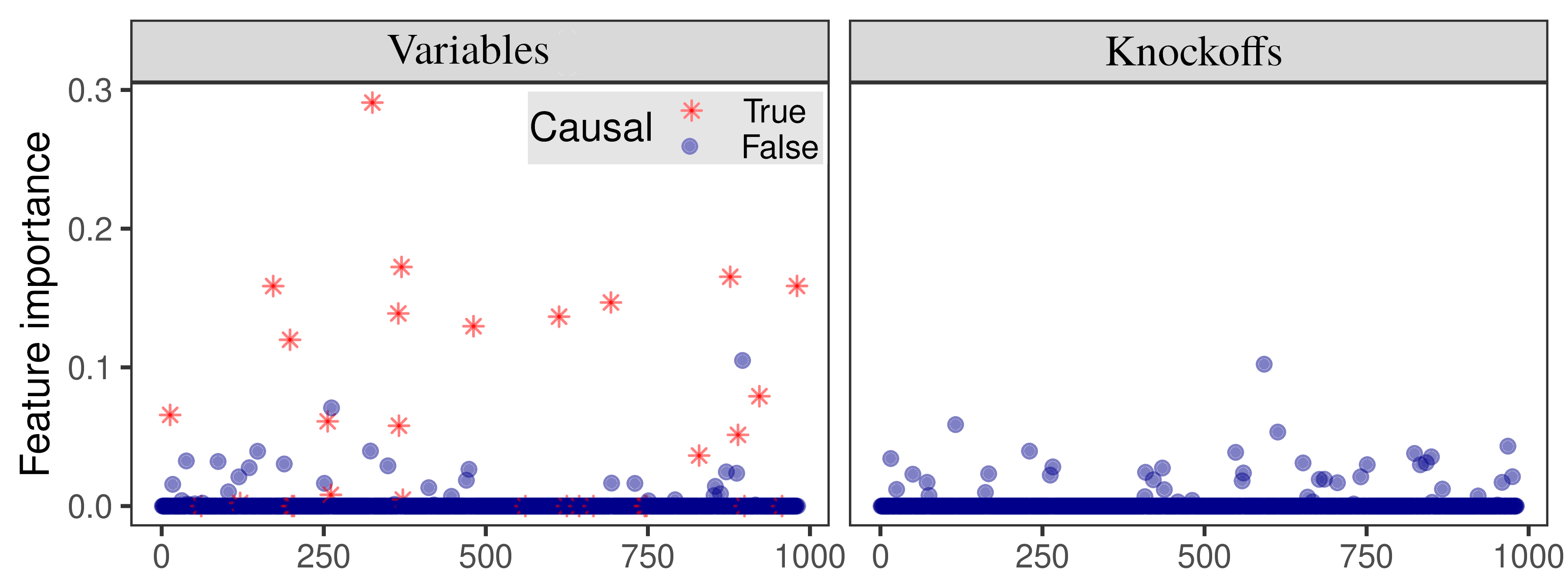
Module 3: test statistics¶
Compute knockoff test statistics for all groups of SNPs, separately for each specified genome partition.
To learn more¶
References:
- False discovery rate control in genome-wide association studies with population structure. M. Sesia, S. Bates, E. Candès, J. Marchini, C. Sabatti. Proceedings of the National Academy of Sciences, 2021. https://doi.org/10.1073/pnas.2105841118
- Multi-resolution localization of causal variants across the genome. M. Sesia, E. Katsevich, S. Bates, E. Candès, C. Sabatti Nature Communications, 2020. https://doi.org/0.1038/s41467-020-14791-2
- Gene hunting with hidden Markov model knockoffs. M. Sesia, C. Sabatti, E. Candès. Biometrika, 2019. https://doi.org/10.1093/biomet/asy033
Other recent works:
Searching for robust associations with a multi-environment knockoff filter. S. Li, M. Sesia, Y. Romano, E. Candès, C. Sabatti. Biometrika, 2021. https://doi.org/10.1093/biomet/asab055
Searching for interactions. Individualized conditional independence testing under model-X with heterogeneous samples and interactions. M. Sesia, T. Sun. (2022, Under review) https://arxiv.org/abs/2205.08653